University Research Council and Carnegie Corporation Postdoctoral Fellowship in Global Climate Change 2012-2015
Exploring genomic drivers of South Africa's floral diversity: polyploid consequences in Helichrysum odoratissimum
Polyploidy (whole genome duplication) is a key facet of plant biodiversity; it plays a prominent role in plant diversification as approximately 15% of speciation events occur due to polyploidy and ~35% plant genera contain polyploid lineages. The contribution of polyploidy to plant diversification may be of significant importance in biodiverse places where climate has been stable for thousands of years, like South Africa. For this project, I explored whether polyploidy has played a role in lineage diversification in the widespread African species, Helichrysum odoratissimum (ever-lasting daisies, "imphepho"). With Prof. Glynis Goodman-Cron, I examined patterns of genetic structuring of diploid and polyploid populations of H. odoratissimum to determine if genome differences have resulted in reduced gene flow among populations. In addition, morphological data indicate that there are two morphotypes present throughout the species' distribution. Our work suggests that genome variation likely played an important role in diversification of Helichrysum lineages.
Exploring genomic drivers of South Africa's floral diversity: polyploid consequences in Helichrysum odoratissimum
Polyploidy (whole genome duplication) is a key facet of plant biodiversity; it plays a prominent role in plant diversification as approximately 15% of speciation events occur due to polyploidy and ~35% plant genera contain polyploid lineages. The contribution of polyploidy to plant diversification may be of significant importance in biodiverse places where climate has been stable for thousands of years, like South Africa. For this project, I explored whether polyploidy has played a role in lineage diversification in the widespread African species, Helichrysum odoratissimum (ever-lasting daisies, "imphepho"). With Prof. Glynis Goodman-Cron, I examined patterns of genetic structuring of diploid and polyploid populations of H. odoratissimum to determine if genome differences have resulted in reduced gene flow among populations. In addition, morphological data indicate that there are two morphotypes present throughout the species' distribution. Our work suggests that genome variation likely played an important role in diversification of Helichrysum lineages.
National Science Foundation Postdoctoral Fellowship in Bioinformatics 2010-2012
As an NSF postdoc, I worked with Dr. Kari Segraves and Dr. Mark Ritchie to better understand the role of ecology (climatic habitats) in polyploid speciation. My postdoctoral research used a modeling intensive approach with comparative statistics to test whether polyploid plants occupy different climatic niche space from their diploid progenitors. I used previously published works that documented the cytogeography of twenty plant species that occurred either in North America or Europe. Surprisingly, the data showed that polyploids were not more likely to occupy new climatic habitats relative to their diploid relatives (Glennon et al. 2014 Ecology Letters). This work suggests that polyploid speciation may not necessarily be driven primarily by ecological divergence, as was previously hypothesized.
As an NSF postdoc, I worked with Dr. Kari Segraves and Dr. Mark Ritchie to better understand the role of ecology (climatic habitats) in polyploid speciation. My postdoctoral research used a modeling intensive approach with comparative statistics to test whether polyploid plants occupy different climatic niche space from their diploid progenitors. I used previously published works that documented the cytogeography of twenty plant species that occurred either in North America or Europe. Surprisingly, the data showed that polyploids were not more likely to occupy new climatic habitats relative to their diploid relatives (Glennon et al. 2014 Ecology Letters). This work suggests that polyploid speciation may not necessarily be driven primarily by ecological divergence, as was previously hypothesized.
Ph.D. research at The George Washington University 2005-2010, supervised by Dr. Sheri Church
My dissertation research sparked my interest in reproductive isolation, speciation, and polyploidy. I studied whether polyploidy enabled closely related to species to hybridize that would not have necessarily hybridized as diploids. I used population genetic approaches to explore gene flow among tetraploid and diploid populations of six species of the native North American plant genus, Houstonia. Here, I found that gene flow among species likely contributed to the increased genetic diversity of tetraploid lineages. In addition, I used a population genetics approach to identify hybridization patterns between a federally endangered plant species, Houstonia montana (2x; Roan Mountain bluet), and its common sister species, Houstonia purpurea (2x and 4x). I also found that tetraploid H. purpurea populations appeared to hybridize regardless of ploidy. This work contributed to the revision of the USFWS's Conservation Plan for the Roan Mountain bluet.
My dissertation research sparked my interest in reproductive isolation, speciation, and polyploidy. I studied whether polyploidy enabled closely related to species to hybridize that would not have necessarily hybridized as diploids. I used population genetic approaches to explore gene flow among tetraploid and diploid populations of six species of the native North American plant genus, Houstonia. Here, I found that gene flow among species likely contributed to the increased genetic diversity of tetraploid lineages. In addition, I used a population genetics approach to identify hybridization patterns between a federally endangered plant species, Houstonia montana (2x; Roan Mountain bluet), and its common sister species, Houstonia purpurea (2x and 4x). I also found that tetraploid H. purpurea populations appeared to hybridize regardless of ploidy. This work contributed to the revision of the USFWS's Conservation Plan for the Roan Mountain bluet.
Undergraduate research in the Hunter Lab at Salisbury University 2002-2005
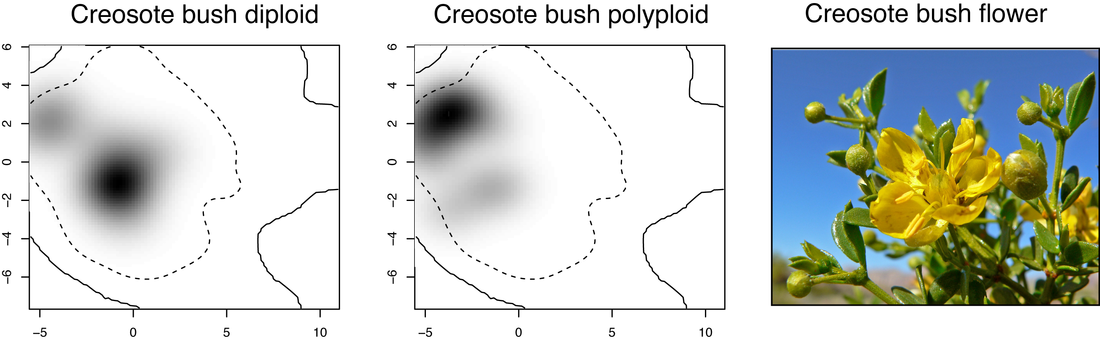
As an undergraduate, I knew I wanted to be involved in finding out new discoveries, encouraging learning, and sharing exciting information with others. This initially landed me in an Early Education degree track at Salisbury University. However, during my sophomore year, I was fortunate to take part in research on polyploidy in Larrea species with Dr Kim Hunter. This transformative experience cemented my passion for learning and, subsequently, plant evolution. While an undergrad volunteer, and later a registered research student, I got a taste for setting up PCRs, running gels, and measuring stomata. I switched my major to Biology with a minor in Chemistry, and never looked back. Dr Hunter provides a supportive environment for her students to present their research (we went to Las Vegas!), seize new opportunities (as an NSF REU at Harvard Forest with Dr Kristina Stinson!), and never stop asking questions (even if it's a similar question in a different group).
As an undergraduate, I knew I wanted to be involved in finding out new discoveries, encouraging learning, and sharing exciting information with others. This initially landed me in an Early Education degree track at Salisbury University. However, during my sophomore year, I was fortunate to take part in research on polyploidy in Larrea species with Dr Kim Hunter. This transformative experience cemented my passion for learning and, subsequently, plant evolution. While an undergrad volunteer, and later a registered research student, I got a taste for setting up PCRs, running gels, and measuring stomata. I switched my major to Biology with a minor in Chemistry, and never looked back. Dr Hunter provides a supportive environment for her students to present their research (we went to Las Vegas!), seize new opportunities (as an NSF REU at Harvard Forest with Dr Kristina Stinson!), and never stop asking questions (even if it's a similar question in a different group).